20:00
Multiomics primer
a discussion about ’omics data analysis and combining modalities
Workshop materials are here:
Goals for this session
Discuss common ’omics issues
Common strategies for adressing them
Simple “integration” methods
Full integration methods (Manon Martin)
Non-goals for today
We can’t give you all the answers today (and we don’t have them!) We hope to give you the right questions to start
- What are my omics datasets really measuring?
- Do I need to transform my data?
- Are my data compositional?
- Are my data sparse?
- What is my goal in combining ’omics data?
- What techniques should I try to connect?
Real Goal - Get you talking about your data challenges
Let’s take a poll
Go to the event on wooclap
Single ’omics Challenges
What did all those ’omics datasets have in common?
What is your research question?
Define your biological question clearly before choosing methods
- Discovery: Find patterns, generate hypotheses
- Prediction: Build models to predict outcomes
- Inference: Test specific biological hypotheses
What are you really measuring?
Understanding your data type matters for analysis choices
- Counts: RNA-seq, 16S - discrete, compositional
- Intensities: Proteomics, metabolomics - continuous, batch effects
- Binary/Categorical: Presence/absence, SNPs
- Each has different assumptions and appropriate methods
Do I need to transform my data?
Raw data often needs transformation for valid statistical analysis
- Log transformation: Stabilizes variance, handles skewness
- Variance stabilizing: DESeq2’s VST, edgeR’s voom
- Scaling/Centering: Makes features comparable
- Batch correction: attempts to remove technical variation
- Check distributions before and after!
Are my data compositional?
Compositional data requires special care
- What is it: Parts of a whole (relative abundance, %)
- The problem: Sum to 1 constraint creates spurious correlations
- Examples: Microbiome, some metabolomics, cell type proportions
- Solutions: CLR transformation, ALR, isometric log-ratio
- Real Solutions Spike-ins, integrating absolute quantification methods
Are my data sparse?
Sparsity (many zeros) affects method choice
- Technical zeros: Below detection limit - consider imputation
- Biological zeros: Truly absent
- Common in: Microbiome, single-cell RNA-seq, proteomics
- Approaches: Zero-inflated models, filtering, imputation
- Trade-off: Filter too much vs. keep noise
Let’s take a poll
Go to the event on wooclap

Case study: Preterm birth
We know the vaginal microbiome is associated with preterm birth
We know preterm birth is often accompanied by inappropriate inflammation
Let’s measure inflammation and the microbiome at the same time, and figure out what our question is later!
Clinical measurments ad metadata
Birth outcome, either “preterm” or “term”
Amsel criteria - a measure of Bacterial vaginosis
- pH
- discharge - thin, watery, greyish
- whiff test - “fishy” polyamine odor
- clue cells - human cells covered in bacteria under microscope
Measuring inflammation
“Inflammation” isn’t one thing - it is an incredibly complex and varied process with cells changing phenotype, proliferating and dying, and signaling each other.
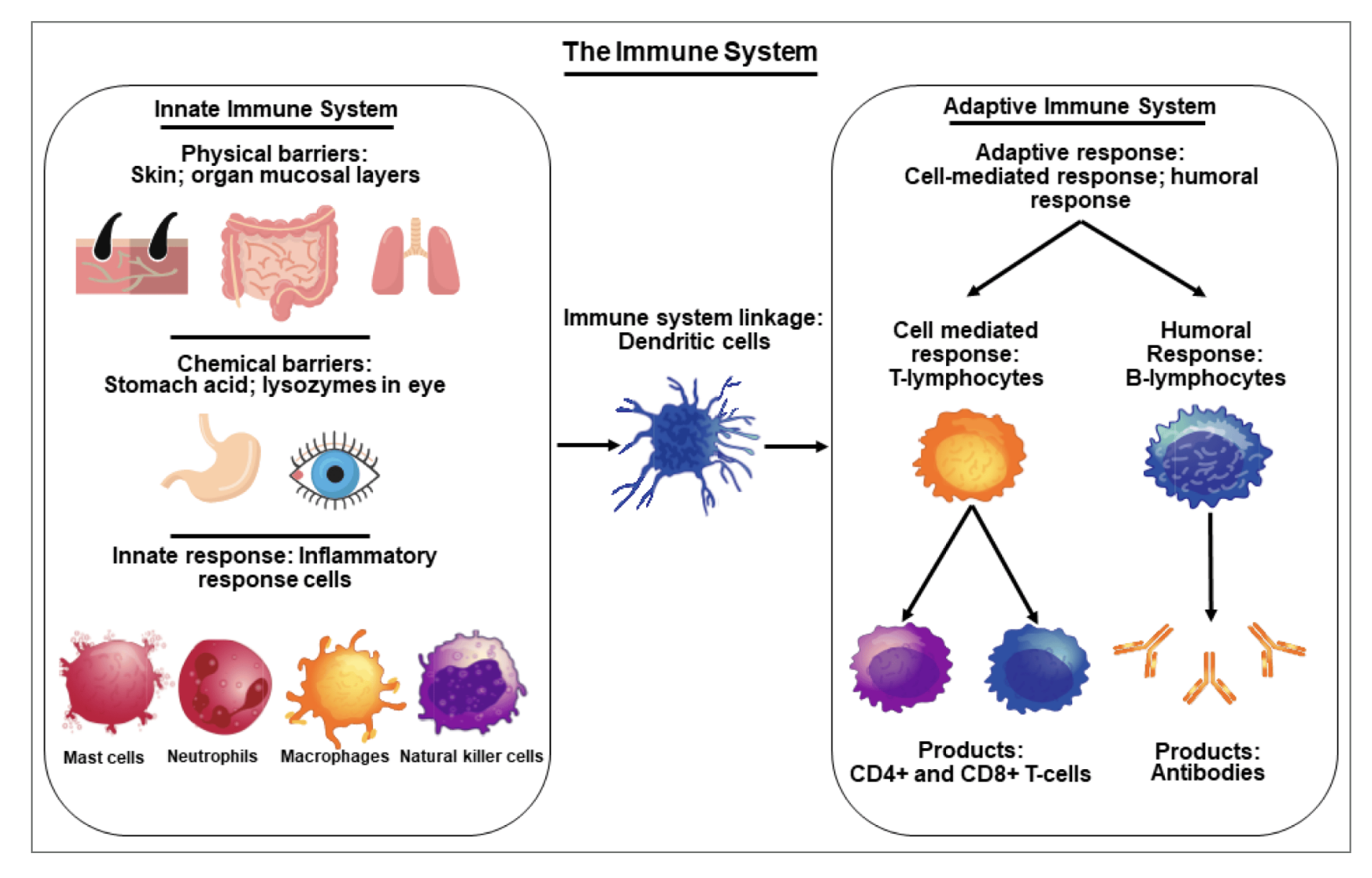
Measuring cytokines
In our case, we’re measuring cytokines which are small secreted peptides that signal immune cells and epithelial cells.
Cytokines are often expressed together and might be redundant. What do we expect when we measure them?
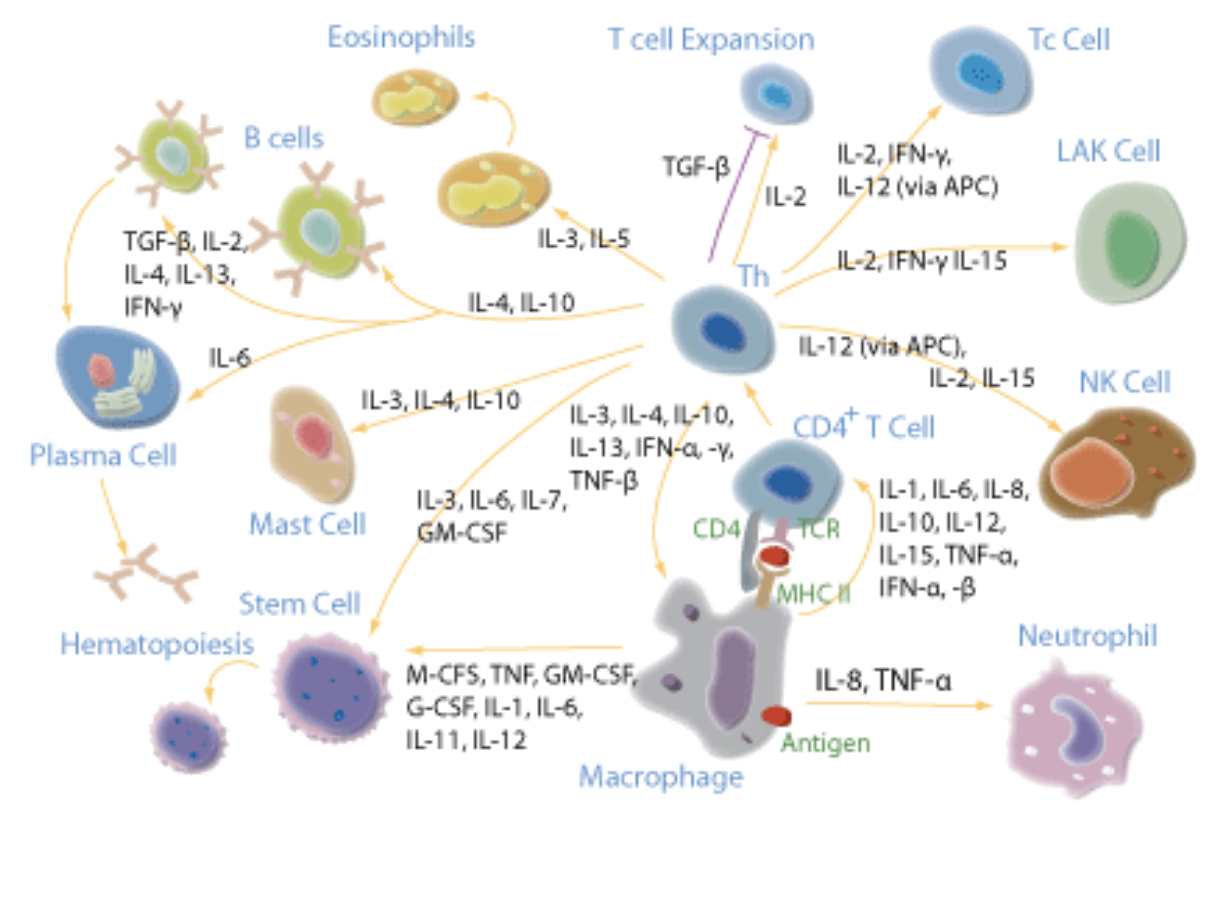
Excalidraw to talk about cytokine measurement
Live coding to show reading, plotting, and transforming cytokine data
Measuring microbiota
What are 3 ways we can measure complex microbial communities?
Excalidraw talking about 16S
Live coding of 16S analysis
- dimensionality reduction
We’ve mastered single ’omics, now let’s combine!
What is your goal in combining ’omics data?
Different goals require different integration strategies
- Improve prediction: Combine features for better models
- Find associations: Correlate features across layers
- Identify pathways: Connect molecular changes
- Understand mechanisms: Causal relationships between layers
- Your goal determines the method!
What techniques should I try to connect?
Start simple, then increase complexity as needed
- Simple: Correlation, differential abundance + metadata
- Intermediate: CCA, PLS, multi-block methods
- Complex: MOFA, DIABLO, network approaches
- Consider: Sample size, data types, computational resources
- Validate: Are results biologically meaningful?
What are your goals for combining datasets?
Get together with a neighbor who doesn’t know your research.
Partner A:
Explain the research question you have
Explain the ’omics data you have / want to generate
Explain how combining the data could be useful (either “metadata” with ‘omics, or multiple high-dimensional datasets’)
Reporting back
Go to the event on wooclap

Reporting back
One brave soul explain your partner’s project and why they want to integrate multiple ’omics datasets
05:00
Methods for Multi-omics integration
Manon Martin, Laura Symul, and Laura Vermeren