NextSeq2000 / NovaSeq Demultiplexing and Data Transfer
- Mount the remote SMB server. You must be on a secure network (MGBWifi or set up a partners vpn)
sudo mount -t cifs //10.212.9.248/KwonLabSequenceData ~/remoteDir -o "username=Sequser-read,password=1234R@g0n"- Make sure you are able to access the SMB via your file explorer.
- Transfer from SMB onto O2 scratch with RSync
rsync -avt ./{sequencing_folder} {hms_id}@transfer.rc.hms.harvard.edu:/n/scratch/users/{first_letter_hms_id}/{hms_id}- Make demultiplexing samplesheet
- bclConvert header
Sample_IDas the unique identifier- index2 as the
- index1 as the
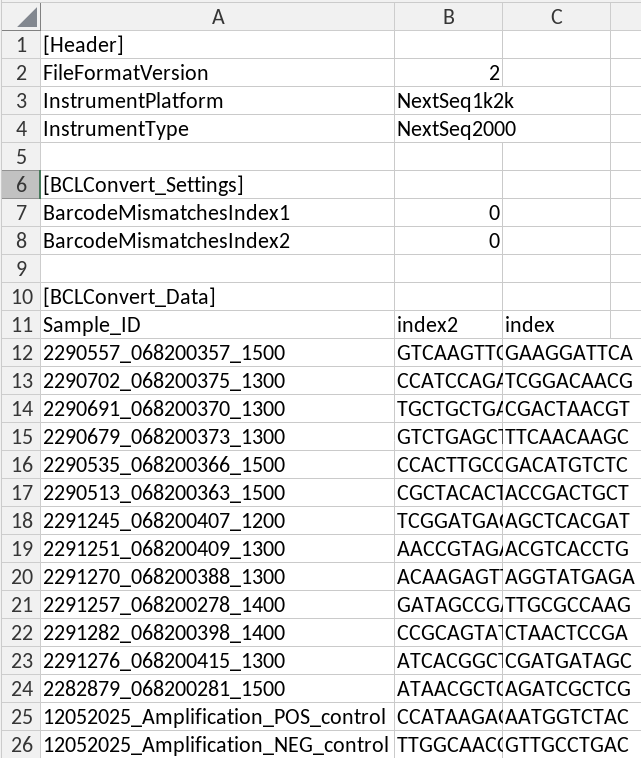
Upload demultiplexing samplesheet to o2 via o2 portal
Validate and upload mapping file with sequencing metadata for dada2 to data1 folder
- Make sure format is consistent with old miseq runs
#SampleIDas the unique identifier- Ensure no special characters in the unique identifier and only (alphanumeric and underscores)
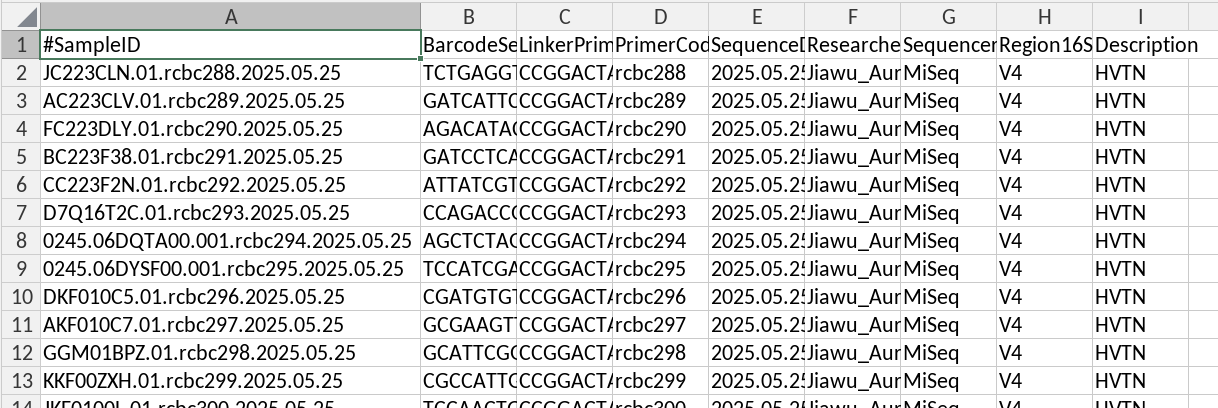
- Run bclConvert
- Ensure you are on an interactive node with at least 96GB of memory
module load bclConvert
bcl-convert --bcl-input-directory . --output-directory fastqs --sample-sheet vibrant_sample_sheet_pool11.csv- Move fastqs to data1 folder
n/groups/kwon/data1/sequencing_run_archive_DO_NOT_EDIT/2025_05_07_NextSeq_V3V4/
├── Demultiplexed
├── Reports
└── vibrant_pool10.csvThe sequencing run is now demultiplexed, in the proper directory on O2, and ready for further processing.